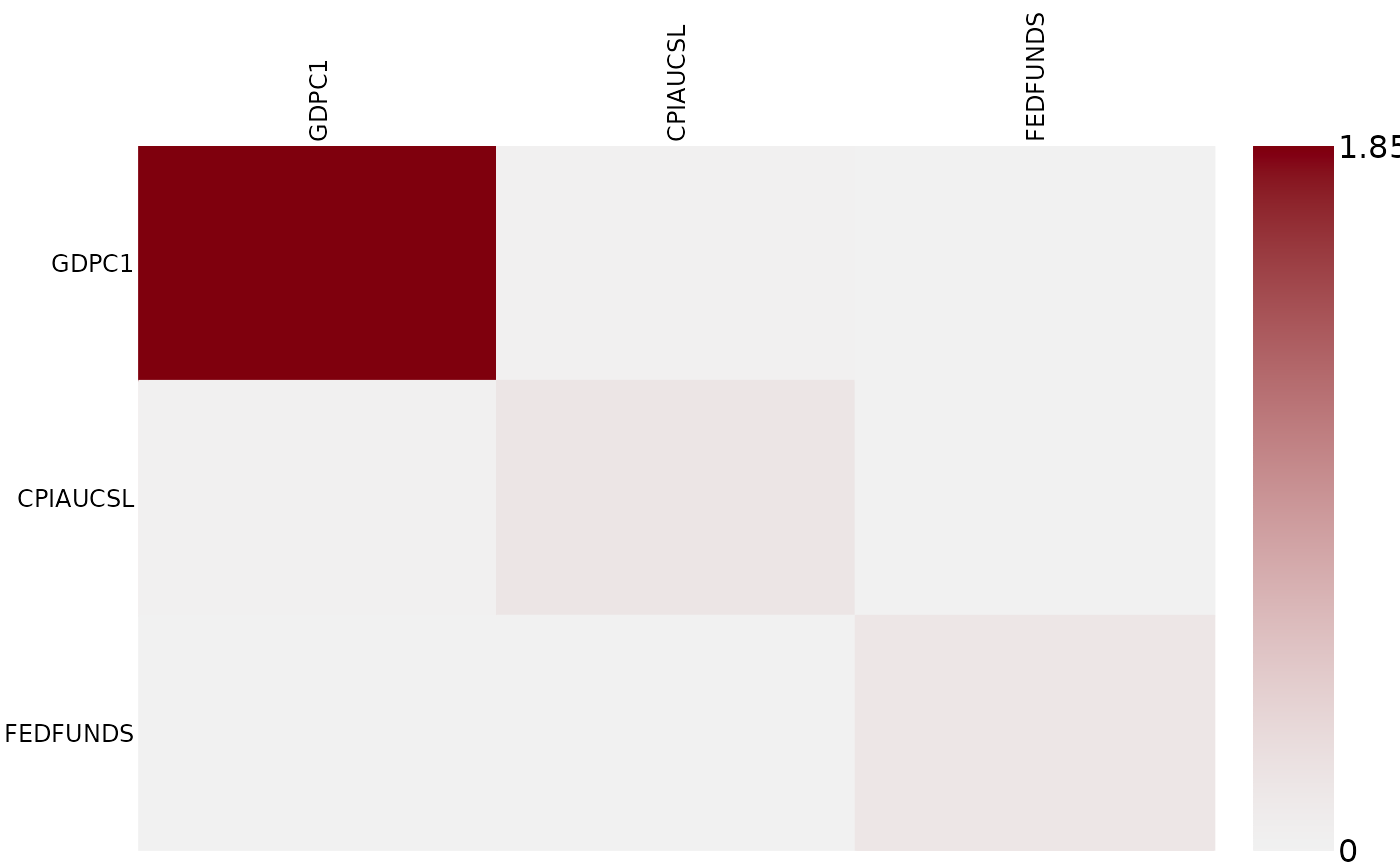posterior_heatmap() generates a heatmap for draws of matrix values
parameters visualizing point wise summaries, such as mean, median, variance,
standard deviation, interquartile range etc. etc.
Usage
posterior_heatmap(
x,
FUN,
...,
transpose = FALSE,
colorbar = TRUE,
colorbar_width = 0.1,
gap_width = 0.25,
xlabels = NULL,
ylabels = NULL,
add_numbers = FALSE,
zlim = NULL,
colspace = NULL,
border_color = NA,
zero_color = NA,
main = "",
detect_lags = TRUE,
cex.axis = 0.75,
cex.colbar = 1,
cex.numbers = 1,
asp = NULL
)Arguments
- x
An array of dimension \(a \times b \times draws\), where \(a \times b\) is the dimension of the parameter to visualize and draws is the number of posterior draws.
- FUN
The summary function to be applied to margins
c(1,2)of x. E.g."median","mean","IQR","sd"or"var".apply(x, 1:2, FUN, ...)must return a matrix!- ...
optional arguments to
FUN.- transpose
logical indicating whether to transpose the matrix or not, i.e. whether to plot an \(a \times b\) or an \(b \times a\) matrix. Default is
FALSE.- colorbar
logical indicating whether to display a colorbar or not. Default is
TRUE.- colorbar_width
numeric. A value between 0 and 1 indicating the proportion of the width of the plot for the colorbar.
- gap_width
numeric. A value between 0 and 1 indicating the width of the gap between the heatmap and the colorbar. The width is computed as
gap_width*colorbar_width.- xlabels
ylabels=NULL, the default, indicates thatcolnames(x)will be displayed.ylabels=""indicates that no ylabels will be displayed.- ylabels
xlabels=NULL, the default, indicates thatrownames(x)will be displayed.xlabels=""indicates that no ylabels are displayed.- add_numbers
logical.
add_numbers=TRUE, the default indicates that the actual values ofsummarywill be displayed.- zlim
numeric vector of length two indicating the minimum and maximum values for which colors should be plotted. By default this range is determined by the maximum of the absolute values of the selected summary.
- colspace
Optional argument indicating the color palette to be used. If not specified,
colorspace::diverge_hcl()will be used, or, ifFUNreturns only positive valuescolorspace::sequential_hcl(). See below for a more detailed description of the default usage.- border_color
The color of the rectangles borders. If not specified no borders will be displayed.
- zero_color
The color of exact zero elements. By default this is not specified and then will depend on
colspace.- main
main title for the plot.
- detect_lags
logical. If
class(x)is "bayesianVARs_coef", thendetect_lags=TRUEwill separate the sub matrices corresponding to the lags with black lines.- cex.axis
The magnification to be used for y-axis annotation relative to the current setting of cex.
- cex.colbar
The magnification to be used for colorbar annotation relative to the current setting of cex.
- cex.numbers
The magnification to be used for the actual values (if
add_numbers=TRUE) relative to the current setting of cex.- asp
aspect ratio.
Examples
# Access a subset of the usmacro_growth dataset
data <- usmacro_growth[,c("GDPC1", "CPIAUCSL", "FEDFUNDS")]
# Estimate a model
mod <- bvar(100*data, sv_keep = "all", quiet = TRUE)
# Extract posterior draws of VAR coefficients
phi_post <- coef(mod)
# Visualize posterior median of VAR coefficients
posterior_heatmap(phi_post, median, detect_lags = TRUE, border_color = rgb(0, 0, 0, alpha = 0.2))
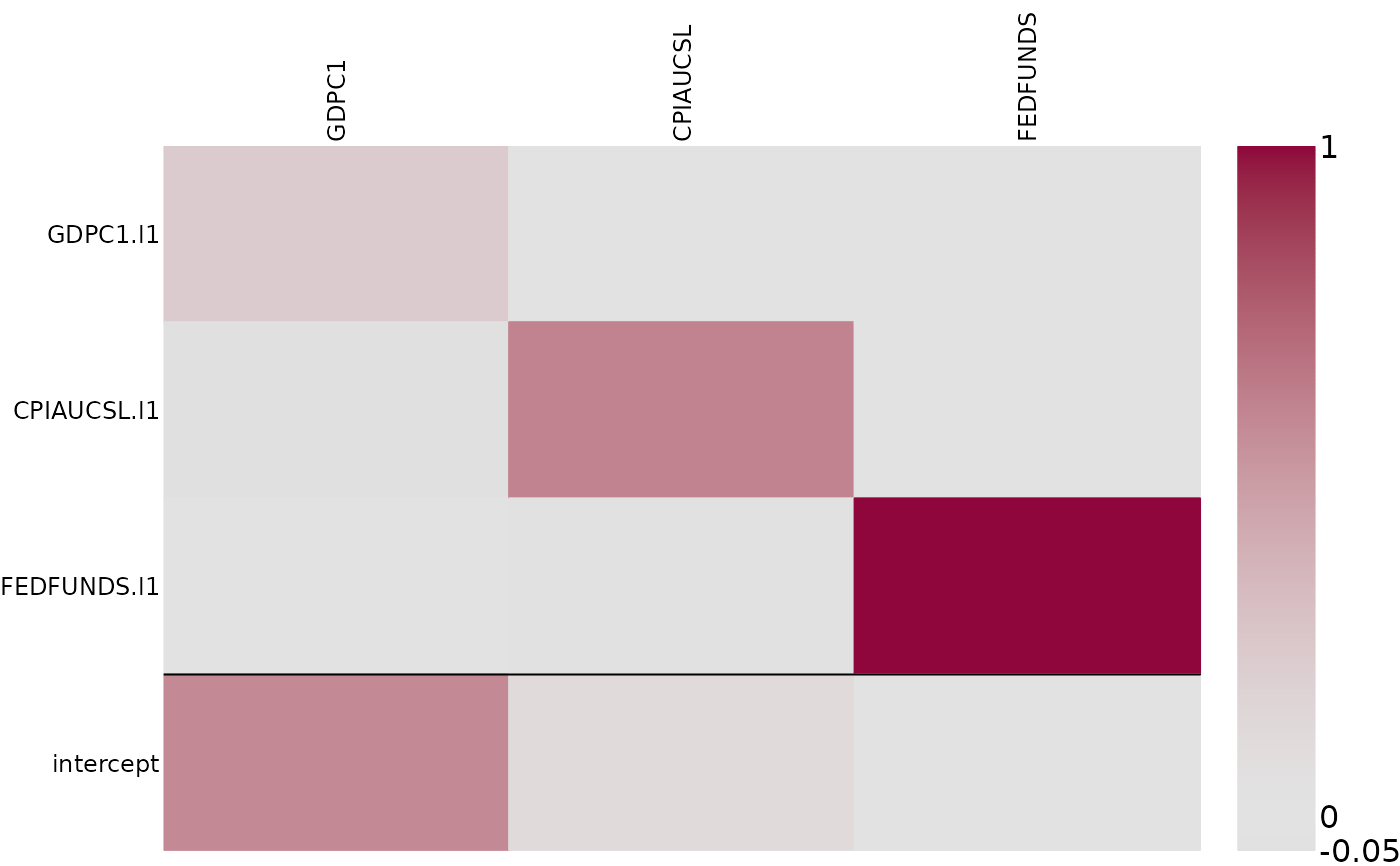 # Extract posterior draws of variance-covariance matrices (for each point in time)
sigma_post <- vcov(mod)
# Visualize posterior interquartile-range of variance-covariance matrix of the first observation
posterior_heatmap(sigma_post[1,,,], IQR)
# Extract posterior draws of variance-covariance matrices (for each point in time)
sigma_post <- vcov(mod)
# Visualize posterior interquartile-range of variance-covariance matrix of the first observation
posterior_heatmap(sigma_post[1,,,], IQR)