Retrieve the structural parameter \(\boldsymbol{B}_0\) samples from an IRF object.
Source:R/irf.R
extractB0.RdRetrieve the structural parameter \(\boldsymbol{B}_0\) samples from an IRF object.
Author
Stefan Haan sthaan@edu.aau.at
Examples
train_data <- 100 * usmacro_growth[,c("GDPC1", "GDPCTPI", "GS1", "M2REAL", "CPIAUCSL")]
prior_sigma <- specify_prior_sigma(train_data, type="cholesky", cholesky_heteroscedastic=FALSE)
#>
#> Since argument 'type' is specified with 'cholesky', all arguments starting with 'factor_' are being ignored.
#>
#> Argument 'cholesky_priorhomoscedastic' not specified. Setting both shape and rate of inverse gamma prior equal to 0.01.
mod <- bvar(train_data, lags=5L, prior_sigma=prior_sigma, quiet=TRUE)
structural_restrictions <- specify_structural_restrictions(
mod,
restrictions_B0=rbind(
c(1 ,NA,0 ,NA,NA),
c(0 ,1 ,0 ,NA,NA),
c(0 ,NA,1 ,NA,NA),
c(0 ,0 ,NA,1 ,NA),
c(0 ,0 ,0 ,0 ,1 )
)
)
irf_structural <- irf(
mod, ahead=8,
structural_restrictions=structural_restrictions
)
B0 <- extractB0(irf_structural)
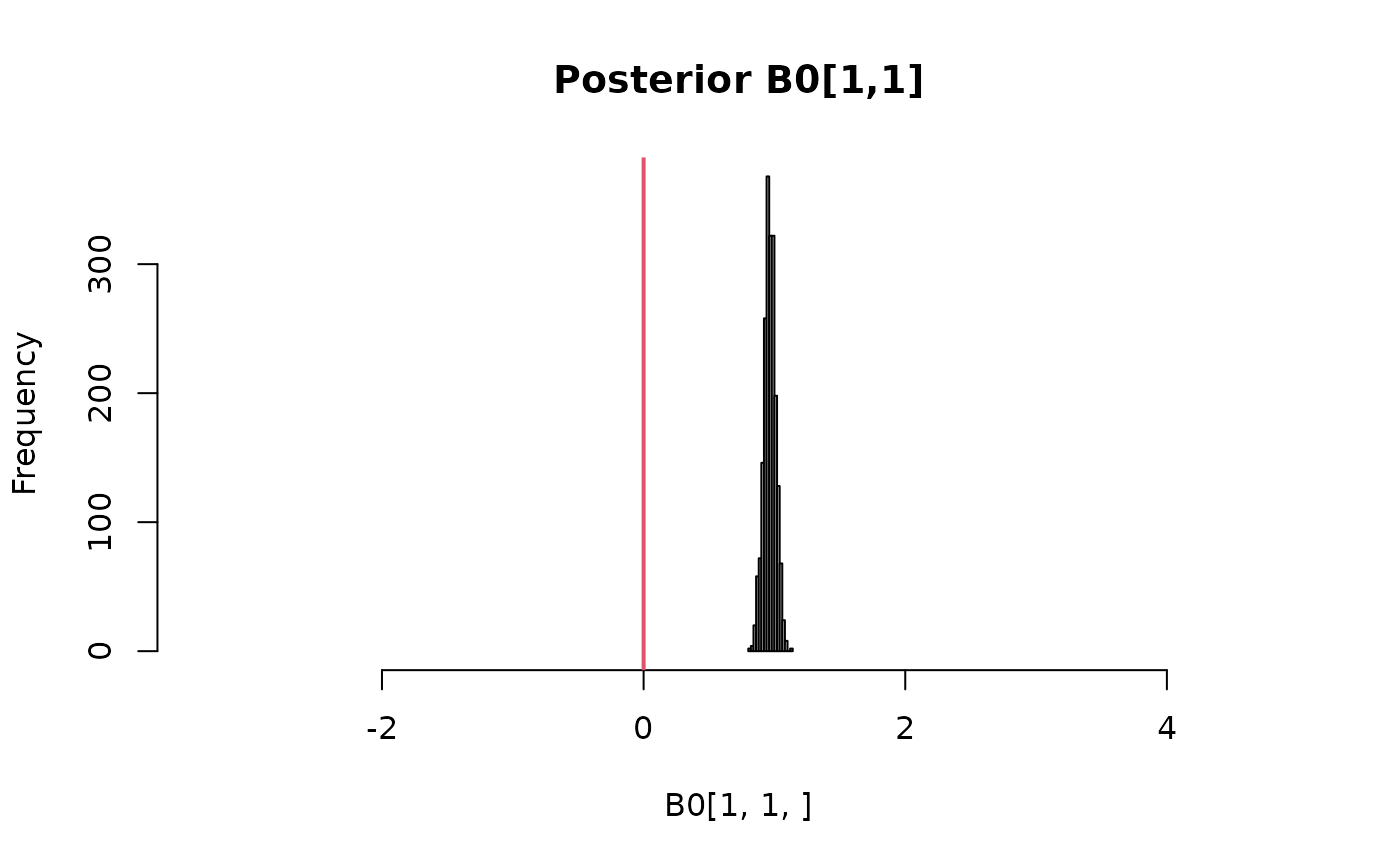
# Visually check that the restriction B0[1,1] >= 0 has been satisfied
hist(
B0[1,1,],
xlim=range(0, B0),
main = paste0("Posterior B0[", 1, ",", 1,"]")
)
abline(v=0, col=2, lwd=2)